The Pitfalls of Spatial Domain Identification
Well-known Machine Learning mistakes can steer Spatial Omics in the wrong direction
Imagine you’ve spent months developing a cutting-edge tool, only to discover that the way everyone measures success might be flawed.
In August 2020, I began my PhD with the idea of working on spatial domains, a concept still in its infancy. At the time, only one method had been published. Fast forward four years, and spatial omics has exploded in popularity, with over 87 methods for identifying spatial domains now available. That’s right. Eighty-seven.
In my last post, I shared how I ditched my fancy plan of using Graph Neural Networks and instead developed a simpler method called CellCharter1. Once it was ready, we needed to see how it compared against other methods, and for that, we turned to existing benchmarks used in the field.
However, we realized that nearly all benchmarks suffered from two of the most common pitfalls in statistics and machine learning.

The Human Cortex Dataset
To understand these pitfalls, let’s first look at the dataset most commonly used for benchmarking. It involves 12 samples of the human cortex, a brain region with six distinct layers, each containing different combinations of cell types.
Researchers used the Visium platform from 10x Genomics to capture gene expression in each spot of the tissue, providing spatially resolved transcriptomics data2. This grid-like dataset links gene expression with physical locations on the tissue, allowing scientists to assign spots to the appropriate cortex layer.

When the authors behind stLearn3, the first spatial domain method, tried to group spots based on their individual gene expression alone, the results were quite messy. The groups would only loosely match with the cortex layers and would not be well-defined regions.
However, when they considered each spot's gene expression along with its neighbors, the core idea behind spatial domains, the groups matched the layers better. Not perfect, but good enough for everyone to say, "Yup, let’s use this to benchmark everything else from now on.”

The ability to obtain spatial domains matching these cortical layers became the foundation for benchmarking almost every spatial domain method that followed.
But here’s the catch—actually, two catches.
Pitfall #1: Ignoring Randomness
All spatial domain methods include an element of randomness. Train the same model twice with identical settings, and you might get slightly different results. This variability isn’t inherently bad, but when it’s ignored during model comparisons, it can mislead us.

So, if a method is evaluated on a single run, it might come out on top just because the universe decided to be in a good mood that day.
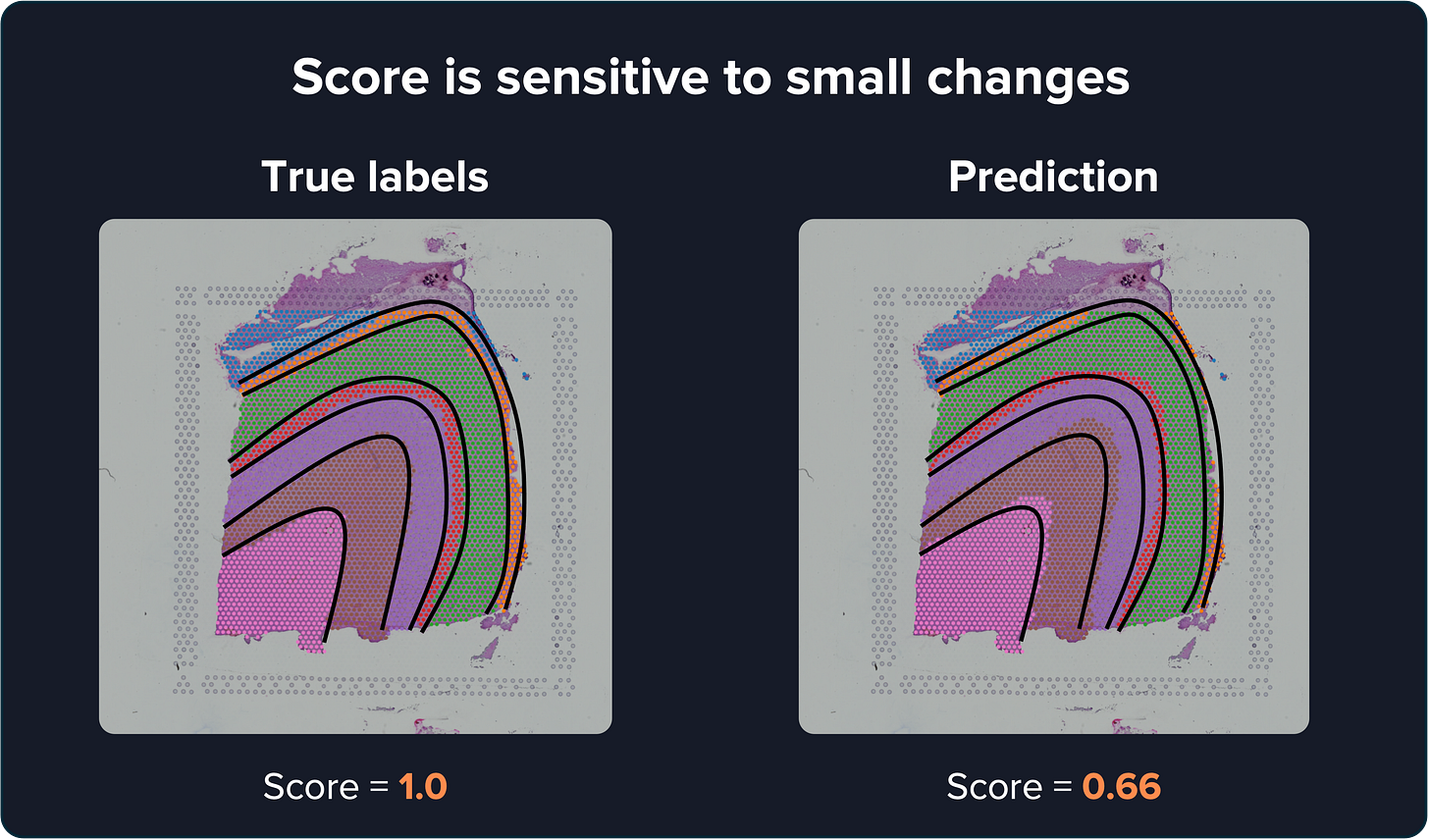
Especially because small differences in the predicted labels can dramatically affect the score. For example, if I take the true labels and shift them by only a few spots, the similarity score drops from 1 to 0.66. That’s a big difference for something so small.
To address this, we had to take a more robust approach: running each model multiple times (we chose 10) and averaging the results.
Is it a pain to run? Yes. Does it make you sleep better at night? Also yes.
Pitfall #2: Overfitting
The second is the quintessential statistical pitfall, the one you learn about in any basic machine learning class and that would be the name of my music band if I had one: training on the test set.
Imagine you’re building a model to predict whether an image is of a cat or a dog. If you evaluate the model using the same dataset it was trained on, the model could just memorize the images and labels. As a result, it would perform well on the training data but fail miserably when given new images it hasn’t seen before.
That’s called overfitting.
At first, you might think, “Hey, this doesn’t apply here. The model doesn’t use labels during training, so no worries, right?“ Wrong.
Overfitting of the (hyper)parameters
The issue appears during parameter selection. Each method has parameters that need tuning, and a common practice here is to find the “best” values by evaluating the model on all 12 cortex samples.
Take BayesSpace4, for example. It has several parameters, such as the number of principal components and highly variable genes (HVGs). What is usually done is to test the score for every combination of parameters. What could go wrong? Well, a lot.
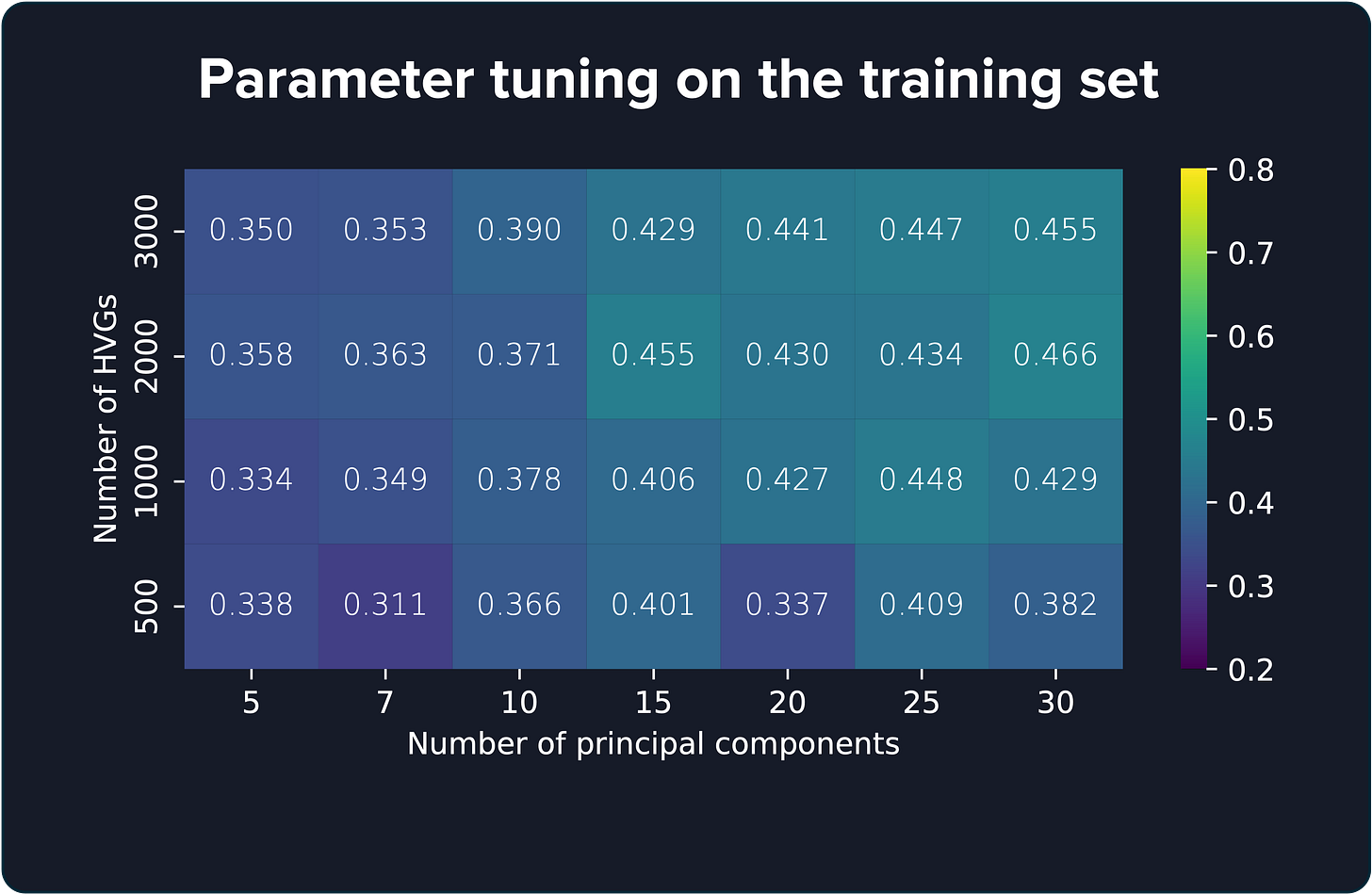
If you test thousands of parameter combinations, eventually you’ll stumble onto a combination that looks amazing. However, given that many attempts, it’s probably a lucky guess. That lucky score won’t hold up if you give the model a new sample it has never seen. It’s classic overfitting.
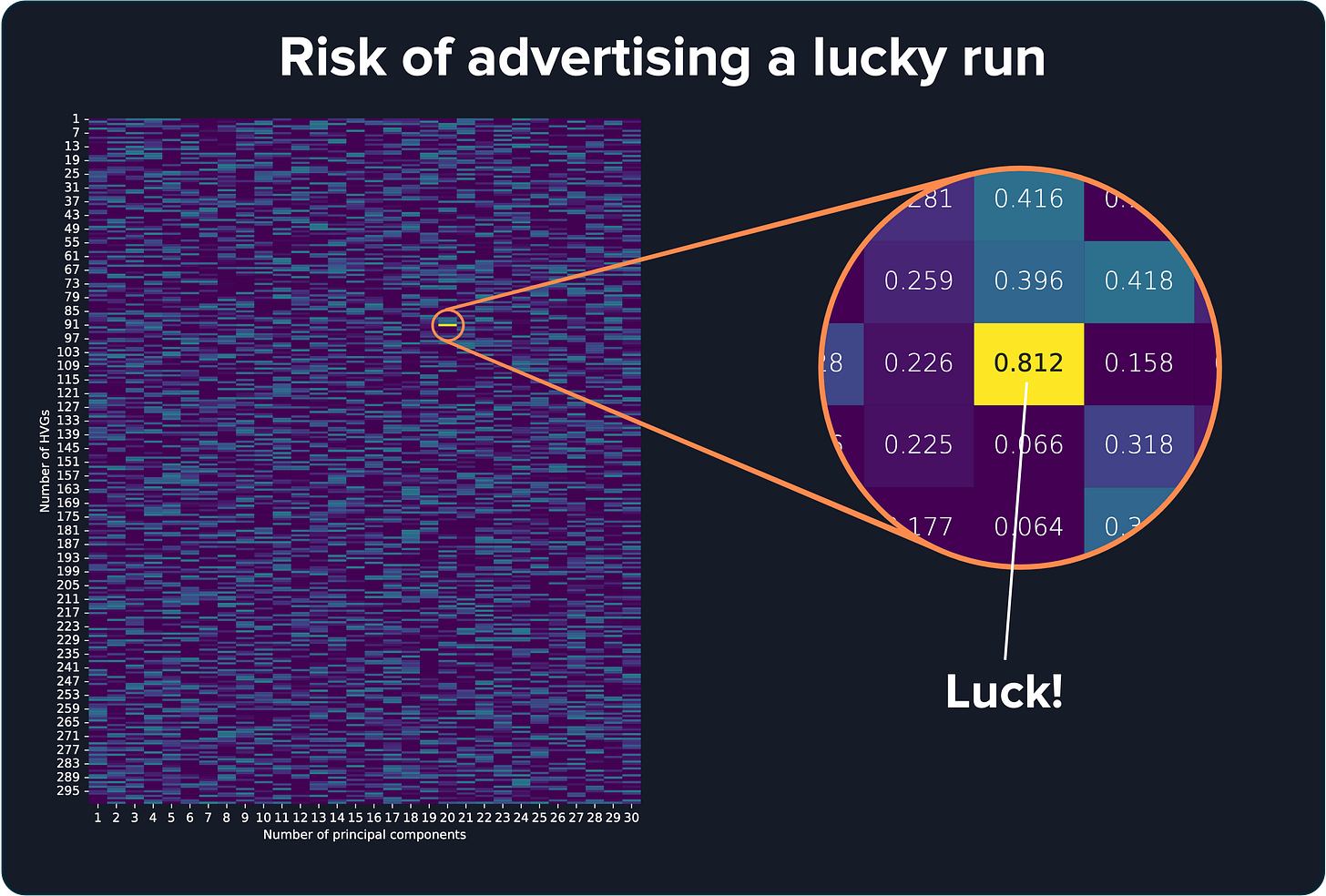
To avoid this, we used a more cautious approach: split the data into training and test sets. We trained on 3 cortex samples, tuned the parameters there, and then tested on the other 9 samples to see if the model could handle new data.

Where do we go from here?
Our changes made the benchmarking process more reliable, but also more complex. We couldn’t evaluate all 87 methods. Testing them all would’ve taken more than a full PhD.
In the end, we focused on the five most commonly used methods, but this raises an important question: how can we efficiently evaluate so many approaches?
The solution is to create a standardized framework that tests multiple methods across diverse, annotated datasets. This would allow researchers to easily plug in their new methods and test them across various tissue types and annotations. After all, if we only test on cortex data, we’ll find the best method for that specific tissue, not the best method overall.
But there is good news! People are working on it:
SpaceHack2.0 started as a hackathon in 2023 and has morphed into a massive project building a framework to test 50 methods on 30 datasets using 20 different metrics. They’re looking for volunteers, by the way! They are also organizing SpaceHack3.0, focusing on best practices for analyzing imaging-based spatial transcriptomics.
Also, there’s a new conference coming up: Modern Benchmarking: Advancing Computational Methods in Molecular Biology in March 2025. If you’re into benchmarking, it’s the place to be.
Bottom line: We need to do better at measuring success in spatial omics. It’s not just about playing with numbers. This stuff could actually help with real-world applications, like, you know, saving lives.
No pressure or anything.

