Breakthrough or incremental progress?
How spatial omics is going to be revolutionary for biology and medicine
The biotech world has witnessed its fair share of overhyped technologies. Some, while potentially groundbreaking, simply didn't live up to the high expectations set for them due to inherent limitations, regulatory barriers, or unforeseen complications.
Today, we stand at the beginning of another potential revolution – spatial omics. Despite the uncertainty, in this post and the next, I am going to show how this technology could mature into an established pillar of biology and medicine.
To explain where spatial omics stands today - on the cusp of medical breakthrough and public anticipation - I will first tell you about the rise and fall of the hype surrounding interferon therapy.
A look back at interferon therapy
It was 1957. At the National Institute for Medical Research, Alick Isaacs, a British virologist, teamed up with Jean Lindenmann, a Swiss post-doc, to study the body's defenses against viral infections. During their studies, they observed that virus-infected cells produced a substance that interfered with the infection of nearby cells by the same or other viruses. Naming this substance was straightforward, given its unique function. They called it interferon, alluding to its capacity to interfere with viral replication. The discovery was groundbreaking.

Initial studies showed promising results, showcasing its powerful antiviral and anticancer properties. The media played a substantial role in elevating the profile of interferon. In the late 1970s and early 1980s, news outlets began to report on the miracle drug that might cure all cancers and combat all viruses. This led to significant investments and competition among biotech firms and pharmaceutical companies to produce and patent interferon therapies.
Over time, as more research was conducted, a deeper understanding of interferon therapy emerged. It became clear that while interferons had therapeutic value in specific contexts, they were not the panacea that early hype suggested. Its application was not universal, and its side effects were not suitable for all patients1.
In essence, the case of interferon therapy underscores the complexities of translating scientific discovery into medical practice. Hype can accelerate interest and funding but can also set unrealistic expectations. As with many medical innovations, the true value and applicability of a therapy often become clearer with time, rigorous research, and real-world clinical experience.
What about spatial omics?
Fast forward to the present, and a technology called spatial omics is capturing a lot of attention in the research community.
Without going into the technical details, spatial omics studies cells in their spatial context within the tissue. As I described in my previous post, it's like studying a city from a detailed map of a city (the tissue sample), whereas before it was only possible to do so from a list of buildings (the list of cells in the tissue).
Is spatial omics heading down the same path as interferon therapy?
Spoiler alert: it’s too early to say. Even though the pace at which technology is evolving seems to increase exponentially, spatial omics only started to emerge as a field in 2016. We are right now during the equivalent of the period between 1957 and the 1970s for interferons.
Early studies that used spatial omics in cancer often reached conclusions such as detecting high heterogeneity of the tumor cells within the same patient (called intra-tumor heterogeneity), which is one of the main causes of therapy failures. However, intra-tumor heterogeneity has been a well-known concept long before spatial omics2, and is detectable using simpler, cheaper technologies.
Yet, there are certain aspects that only spatial omics can capture.
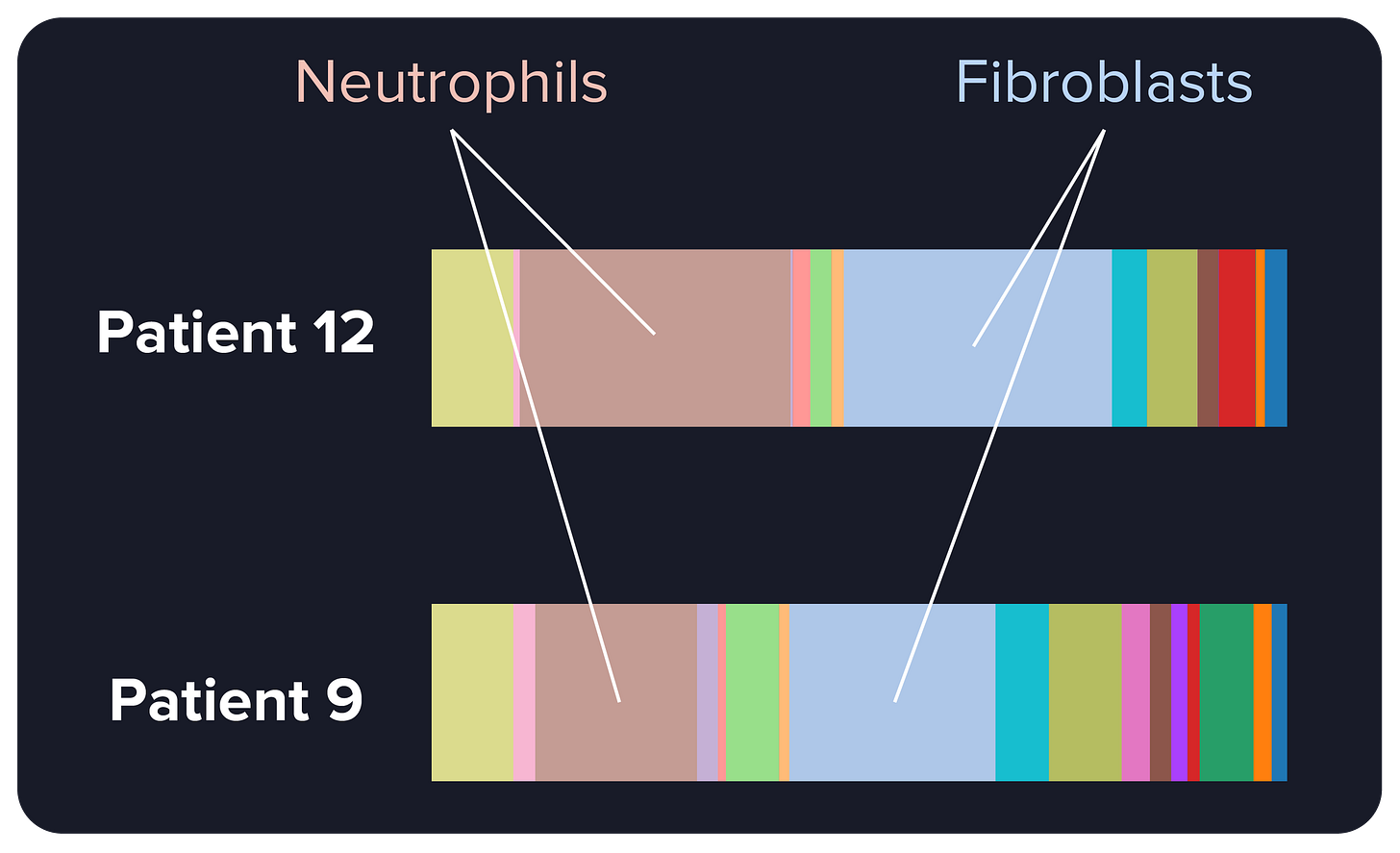
In our study on CellCharter3, a computational method to find recurring communities of cells from spatial omics data, we observed something intriguing. Some lung cancer patients had similar proportions of non-tumor cell types, with a high proportion of cells called neutrophils and fibroblasts.
Through the lens of non-spatial technology, one might think that these patients are very similar, in the sense that their tumor cells are placed in very similar microenvironments.
But once we looked at how those neutrophils and fibroblasts are spatially arranged, a totally different picture came up. These two cell types were intermixed in Patient 12 but physically separated in Patient 9. In these two patients, the cellular communities around the tumor, and the interactions between cells in those communities, are radically different.
Cell interactions are becoming increasingly relevant for cancer therapies, especially immunotherapy, so these new observations may be the key to understanding why a therapy works or fails for a certain patient.
We still don’t have data on whether these differences impact the outcomes of therapies. As I said, we are still in the infancy period of this technology, and having clinical information for these patients is rare. It will take years to prove, or disprove, these observations, but we are finally reaching the equivalent of the 1970s and 1980s for interferon. Hopefully, the research will lead to different and more promising conclusions.



As someone who is beginning to work with spatial omics, I enjoyed reading your viewpoint on the technology. Looking forward to your next article.