Welcome to Batch Effect
A blog on the new frontiers of Computational Biology for cancer research
There are only two hard things in Computer Science: cache invalidation and naming things. - Phil Karlton
Choosing a name for this blog took longer than I had anticipated. I knew it would focus on topics related to Computational Biology and Cancer. It would also include my other academic interests like the analysis of spatial omics data. Don’t worry if you don’t know what spatial omics is, you’ll have plenty of chances to hear about it in future posts.
The challenge was finding a name that would connect the biological world of cells, tissues, and experimental protocols to the computational world of models, metrics, and analyses.
In computational biology, batch effects appear when comparing two biological samples that have been processed differently. Even the smallest difference in the experiment can cause changes in the data that are hard to remove because they often overlap with true biological variation. Let’s see an example.
Degradation or downregulation?
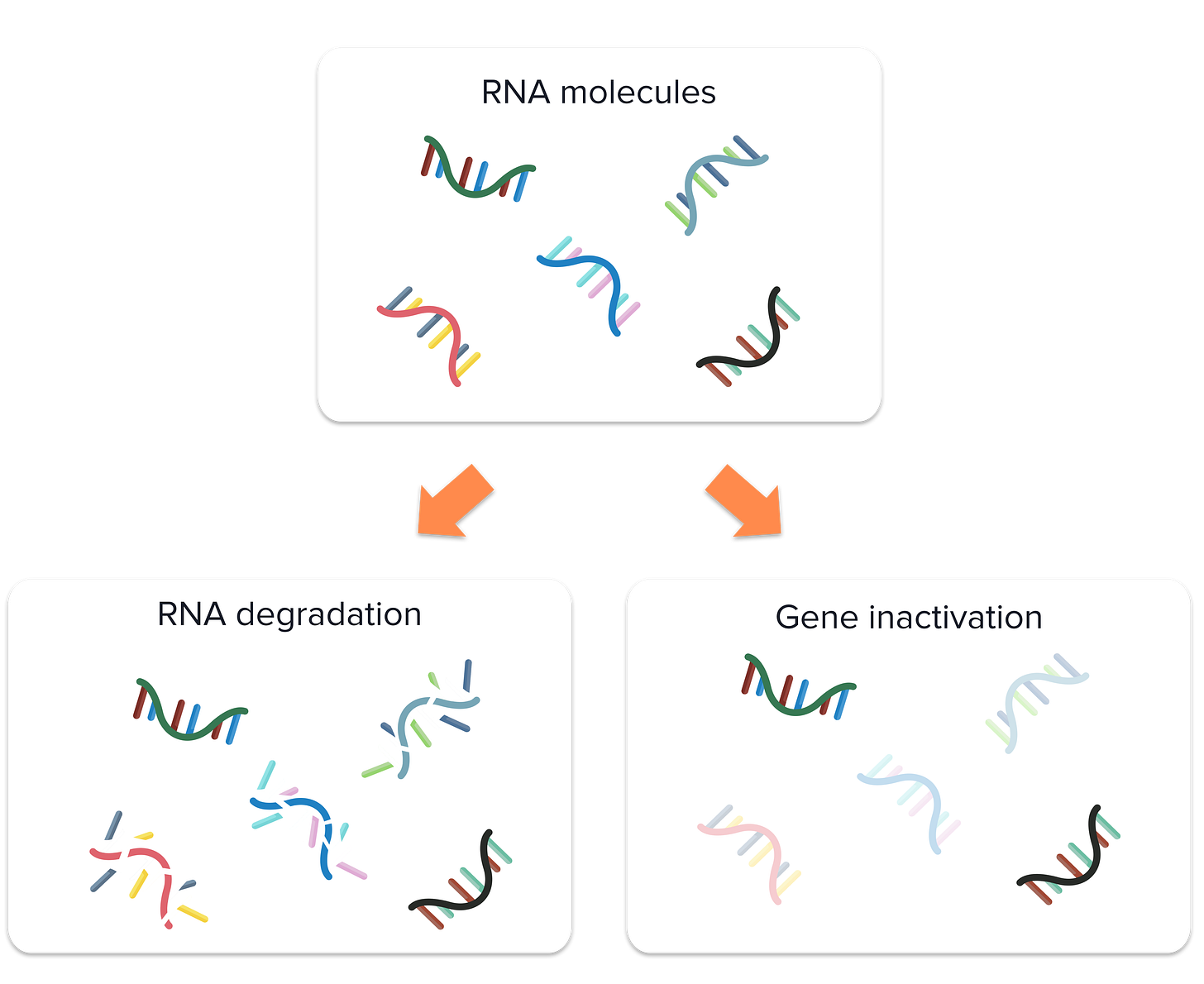
Imagine two samples for which we want to do sequence the RNA: one from a healthy individual and another from patient with a certain disease. Due to logistics, the samples from the healthy individual are processed immediately after collection, while the other one is left at room temperature for a few hours before processing.
Over time, RNA can degrade, especially when exposed to room temperature. This degradation can decrease the abundance of certain RNA molecules. Not all RNAs may degrade at the same rate because some may me less stable than others. As a result, when researchers sequenced the RNA of these samples, the data might indicate decreased levels for specific transcripts in the disease sample compared to the healthy one.
Now, suppose this particular disease inherently causes the inactivation of certain genes. Then, it will be complicated to disentangle whether a gene is expressed at lower levels because of true biological variation or because of the batch effect from RNA degradation.
Why Batch Effect?
Batch effects are effects that every computational biologist will encounter at some point in their career. They are a reminder to young scientists like me, pulling us back from an idealized perception of how science and experiments work.
Batch effects remind us that data, especially biological data, is messy, complex, and inherently noisy. They represent the dance between nature and technology, between the raw truth of biology and the structured logic of computation.
That’s why I chose “the Batch Effect”.
Through Batch Effect, I want to navigate the complexities of cancer research, explore the promising field of spatial omics, and discuss the computational strategies that are pushing the boundaries of what we know about biology.




The problem that batch effects are typically confounded with underlying biology is also something that's bothered me for years. It's especially problematic that most batch effect correction methods (eg the ones that try to match distributions using the maximum mean discrepancy) don't adjust for such confounding. And methods like ComBat that allow you to include confounders are quite limited, because they assume that (1) the effect of the confounder(s) on the features is linear, and (2) that you have access to the confounding variable value on all samples, including new test samples. The latter assumption is especially problematic, because often the confounder is in fact the output variable that we want to predict on new test samples.
I've been working on this as a hobby side project for the last few years, and recently published a paper and released software that addresses this. The basic idea is that you train conditional generative models for the features given the confounder(s), then match the conditional distributions, instead of matching the marginal distributions of the features. Here's a link: https://openreview.net/pdf?id=GSp2WC7q0r . Apologies for shilling my work on your Substack! :)