The giant wakes up: Illumina goes spatial
Predictions on the upcoming Illumina spatial transcriptomics technology
For decades, Illumina has been the name in DNA sequencing.
Many researchers showed that it could become an important player in spatial transcriptomics as well. However, it seemed that Illumina never wanted to jump on this opportunity.
Until now.
In this blog post, I break down everything we know so far about the upcoming Illumina spatial transcriptomics technology:
How Illumina’s technology, originally built for DNA sequencing, can be cleverly adapted for spatial transcriptomics.
A detailed comparison between Illumina’s upcoming product and Visium HD, including a prediction on the actual cost.
The practical implications of Illumina’s promises for research.
The Illumina sequencer
Sequencers determine which DNA or RNA molecules are in a biological sample. These samples often contain a mixture of cells from tissues or fluids, and sequencing helps identify which genes are active, detect mutations, or track pathogens like viruses.
They read the nucleotides (A, C, G, T) of a molecule one by one, but this operation is run for all molecules in parallel.
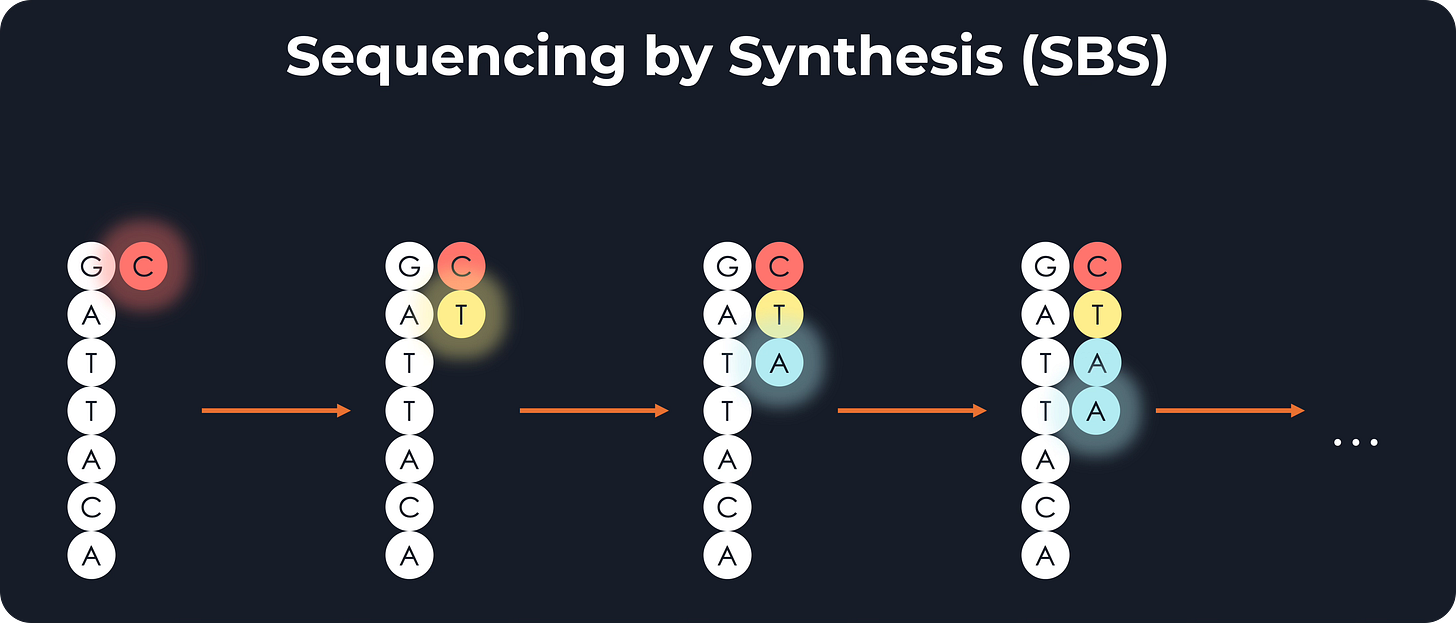
Sequencing
Imagine you have a molecule with the sequence GATTACA. An Illumina sequencer can read it because it has nucleotides floating around that are bound to a specific fluorescent color. As an example, A might be blue, C=red, G=green, and T=yellow. Then, these nucleotides would sequentially bind to their complementary ones in the molecule.
The first nucleotide is a G, so its complementary C would bind, emitting red light. The second is A, so a T would bind, emitting yellow, and so on. By recording the sequence of colors, the machine can deduce the sequence of nucleotides.
However, there may be billions of DNA molecules in a sample. So how do you read them all at once?
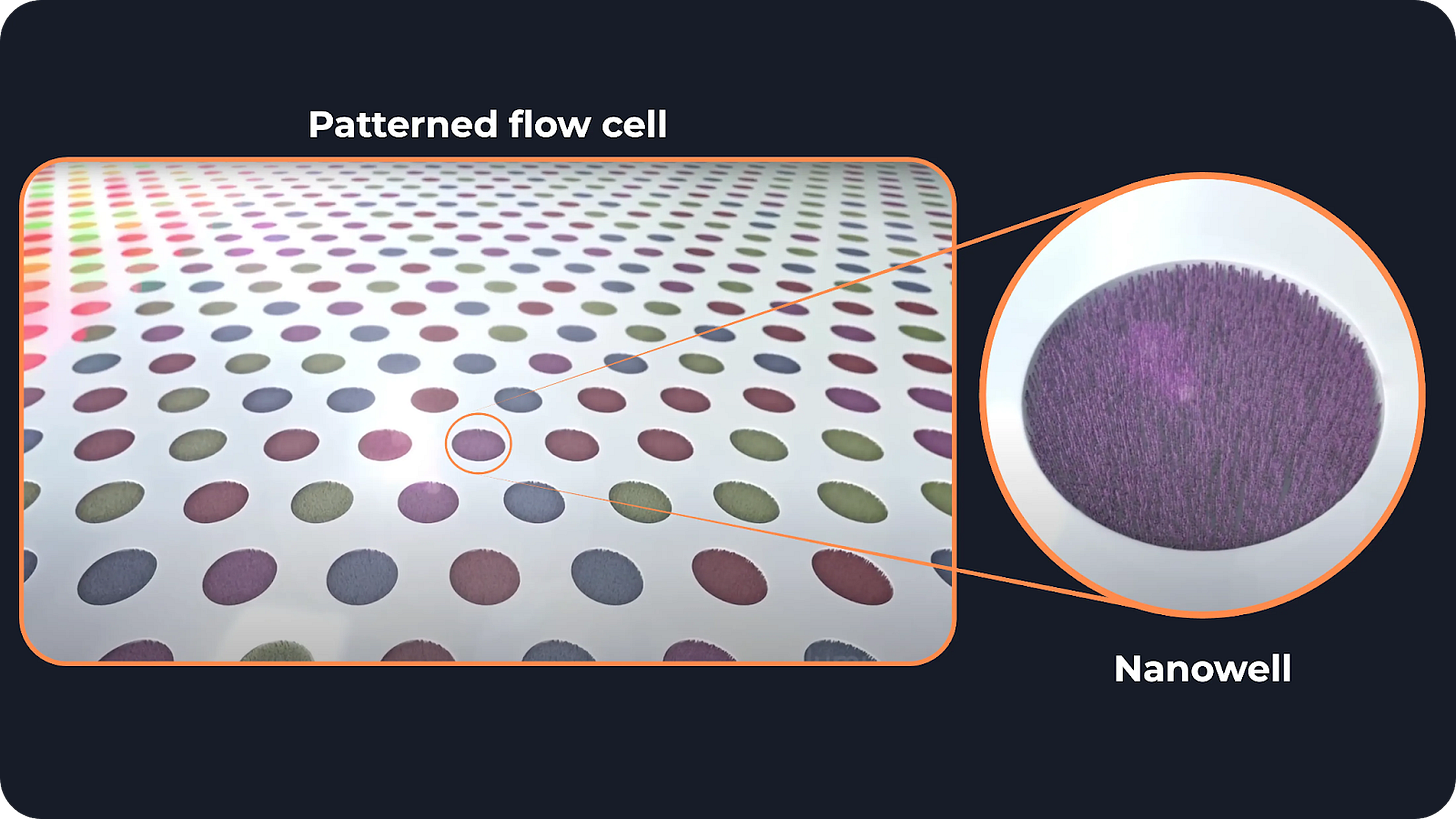
Patterned flow cells
At the heart of Illumina’s approach is its patterned flow cell technology. Think of these flow cells as a grid of microscopic wells, spaced just 0.6 micrometers apart. As a reference, a human cell has a size of 5–20 micrometers.
DNA is extracted from the sample and spread over the flow cell. When a DNA molecule ends up in a well, it starts replicating, forming a cluster of identical copies. The replication happens so fast that there is not time for another molecule to enter the well, so every well contains copies of only one molecule.
And since each cluster consists of many identical DNA molecules, the fluorescence during sequencing is much stronger and easier to detect. This makes the signal way stronger and easier to spot.
Repurposing the grid for Spatial Transcriptomics
A grid of spots, where every spot has a huge amount of the same barcode. Does it remind you of another technology?
This core Illumina technology can be adapted, almost perfectly, for spatial transcriptomics. In 2024, researchers from Max Delbrück Center published Open-ST, where they hacked the flow cell into a spatial transcriptomics platform.
Here’s how:
Artificial barcodes: Instead of sequencing sample DNA, you generate synthetic barcoded DNA molecules that land in the wells of the flow cell.
Barcode mapping: Run a preliminary sequencing to map where each barcode landed.
Tissue transfer: A thin tissue section is placed over the flow cell. RNA from the tissue binds to the barcodes below.
Reverse transcription and sequencing: The RNA is converted into DNA linked to spatial barcodes, then sequenced.
Spatially resolved data: Combining barcode identity with pre-mapped positions creates a high-resolution gene expression map. Other similar examples include Pixel-seq and Seq-scope.
How does it compare to other technologies?
Although Illumina sequencers have been repurposed for spatial transcriptomics since 2022, Illumina’s official product isn’t launching until 2026. Why such a delay?
My hypothesis is that Illumina is not just repurposing flow cells but leveraging its expertise to develop a product more akin to Visium HD. In fact, Illumina’s product doesn’t have gaps between spots, just like Visium HD. This was one of the reasons for 10x Genomics to sue Illumina for patent infringement.
So let’s compare them.
Sample area
Illumina uses a 50×15 mm (7.5 cm²) slide versus a slide of two 6.5×6.5 mm capture areas in Visium HD (0.4 cm² total).
While large samples are appealing for studying tissue heterogeneity, excessively large sample areas may not always be practical. Researchers will have to fit multiple samples into the same slide, at the risk of tissue damage or lower quality of the results.
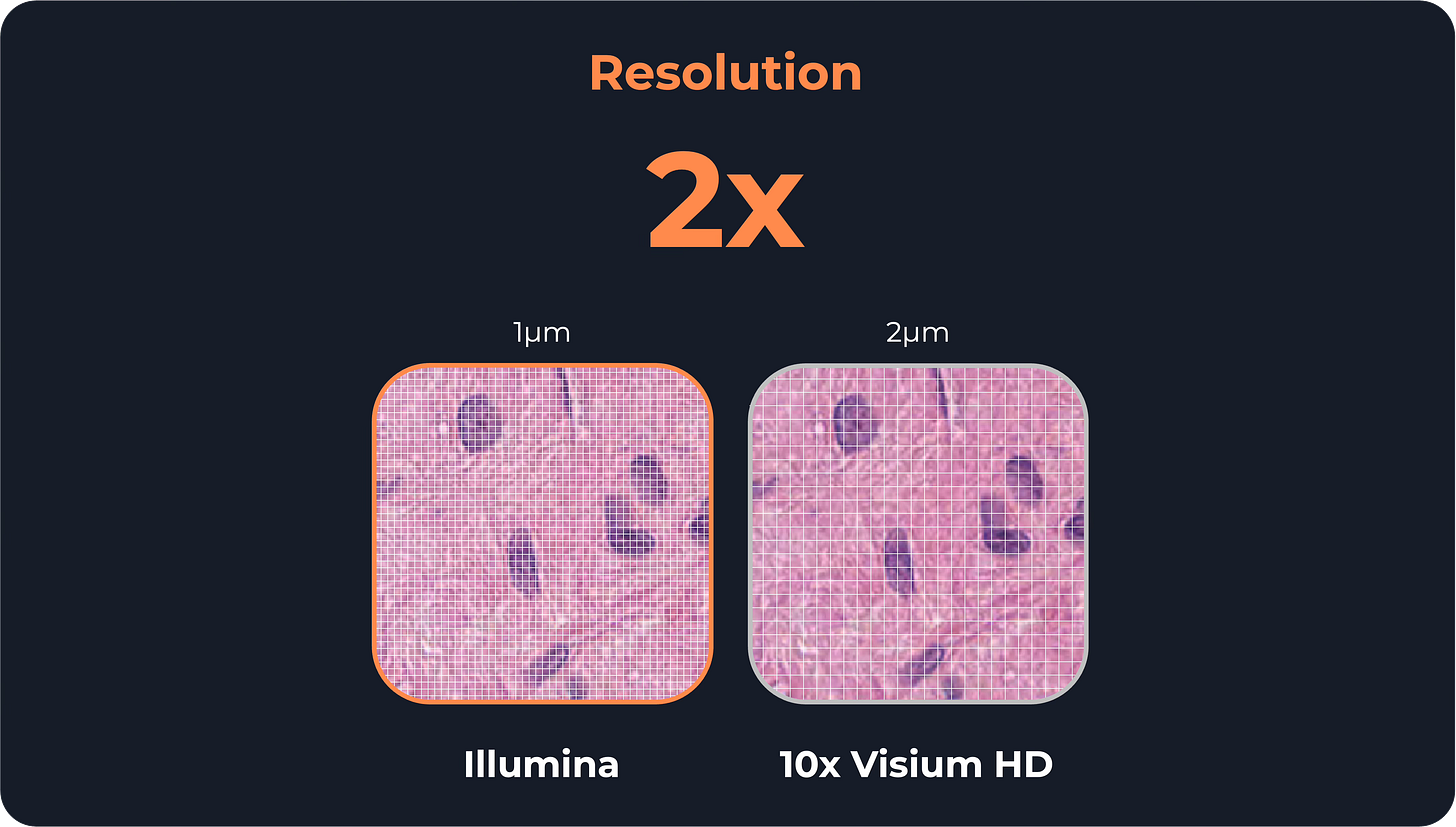
Resolution and sensitivity
Illumina claims 1 μm resolution, twice as sharp as Visium HD’s 2 μm
Once you go below single-cell resolution, a 1 μm difference doesn’t make a big difference. That’s because when RNA molecules move vertically from the tissue to the barcoded spots, they also tend to move sideways and can end up 10-20 micrometers away from where they started (a process called lateral diffusion).
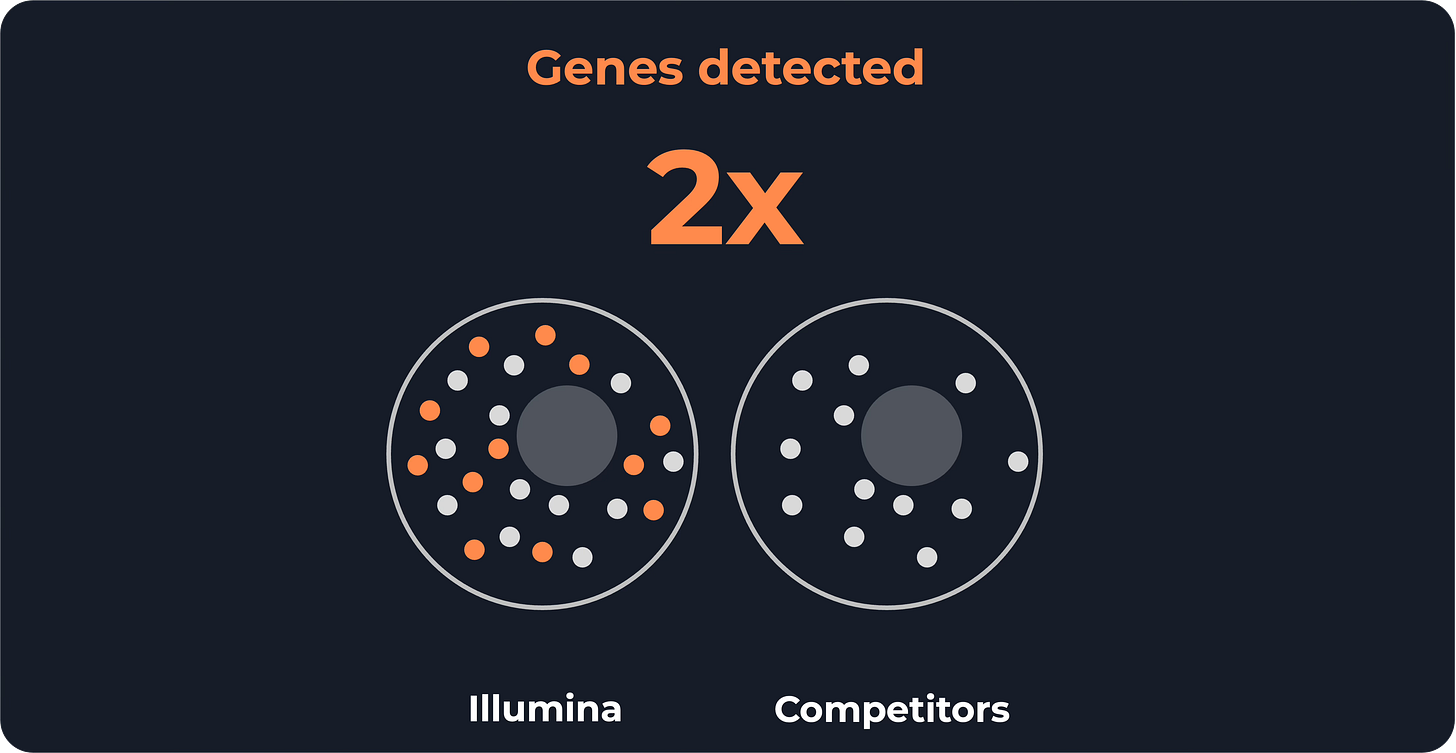
However, the real gain is barcode density: Illumina increased it by 4x. That boosts sensitivity, given that current Visium HD experiments give an average of 200-400 transcripts per cell, compared to the 4,000-10,000 in a scRNA-seq experiment.
Indeed, Illumina claims to detect 2× more genes than competitors.
Cost and sensitivity
Here’s where things get interesting, and where Illumina’s marketing claims need a reality check.
Illumina also claims to lower the cost per tissue area by 4x, compared to other sequencing-based spatial transcriptomics technologies.
But, as Prof. Krisztian Homicsko noted, it’s unclear whether sequencing costs are included in the comparison. My belief is that the estimates do not include the cost of sequencing, but I would be happy to be contradicted.
Illumina people are reading this post, you know how to find me :)
Furthermore, Illumina’s product is 4x cheaper if we manage to cover the whole slide. But, as I mentioned before, you would need amazing Tetris skills to fit 5-10 tissue samples in a single slide.
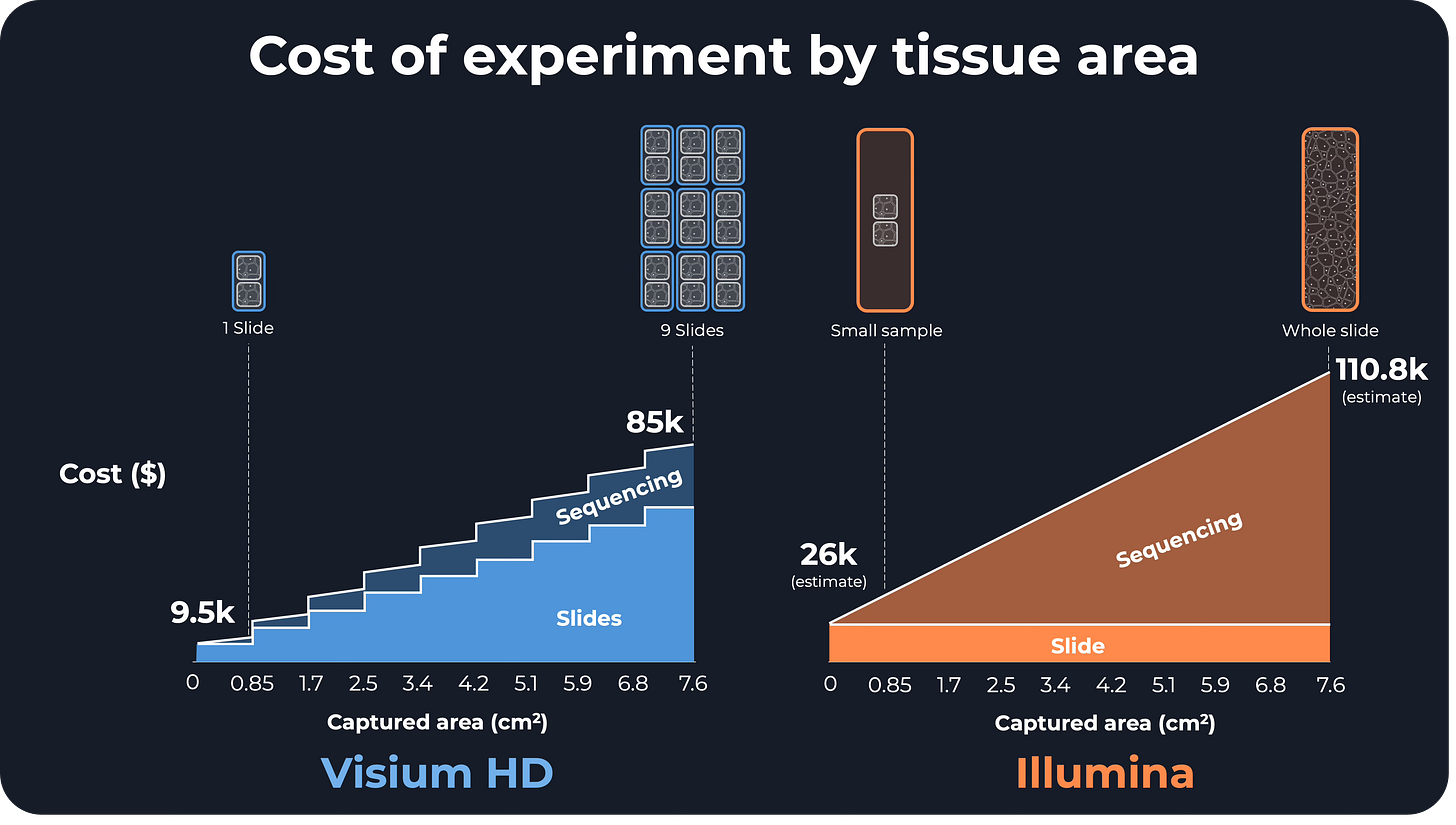
To better understand the implications, I did some back-of-the-envelope estimations of the cost of an Illumina experiment with respect to the size of the tissue sample, taking an example cost for Visium HD from the Emory University Genomics Core facility website.
DISCLAIMER: All these results are estimates, as the pricing has not been released by Illumina yet.
For a small sample with an area of 0.8 cm² (the equivalent of 2 Visium HD capture areas):
Visium HD: $9.5k ($6.8k slide + $2.7k sequencing)
Illumina: $26k ($15.3k slide + $10.7k sequencing)
Illumina is 2.7x more expensive than Visium HD.
For a big sample with an area of 7.2 cm² (18 Visium HD capture areas, which is almost a full Illumina capture area):
Visium HD: $85.1k ($61.2k slide + $23.9k sequencing)
Illumina: $110.8k ($15.3k slide + $95.5k sequencing)
Illumina is still more expensive (1.3x)!
However, most of the cost goes to sequencing rather than the slide cost (86% for Illumina vs 28% for Visium HD), and more sequencing means higher sensitivity.
What this means for research
Thanks to their experience in designing flow cells, Illumina can pack in way more barcodes than the competition. This translates to better sensitivity and more genes detected.
The cost story is more nuanced than Illumina’s marketing suggests.
For small samples, stick with Visium HD, and if you want to have high sensitivity, consider running scRNA-seq on an adjacent slide. The cost will still be lower than running Illumina.
For big samples, or if you can somehow fit many samples into one slide, Illumina could be the better deal. Also because, in the case of Visium HD, you will have to pay for the people and time to run nine experiments.
That “4x cost reduction” only works if you can fill that entire large slide, which is easier said than done.
But there are also other practical considerations that could impact adoption:
We’re still looking at promises, not proof. Differences in sample preparation, processing time, and throughput between platforms like Illumina and Visium HD could influence their efficiency and cost-effectiveness. Until independent researchers publish datasets using Illumina’s platform, we’re looking at promises, not proven performance.
Experience with the details matters. Companies like 10x Genomics and Nanostring/Bruker invested heavily in experimental and computational improvements to enhance cell segmentation, one of the biggest challenges in spatial transcriptomics. If you can’t tell where a cell is, even the fanciest gene detection doesn’t help much.
Is sensitivity the most important metric? Apparently, yes. Is it worth the cost? It depends. But this will be a topic for another blog post :)
Illumina’s entry won’t revolutionize spatial transcriptomics overnight, but it adds serious competition to a field that could use more options, and that’s good news for researchers everywhere.